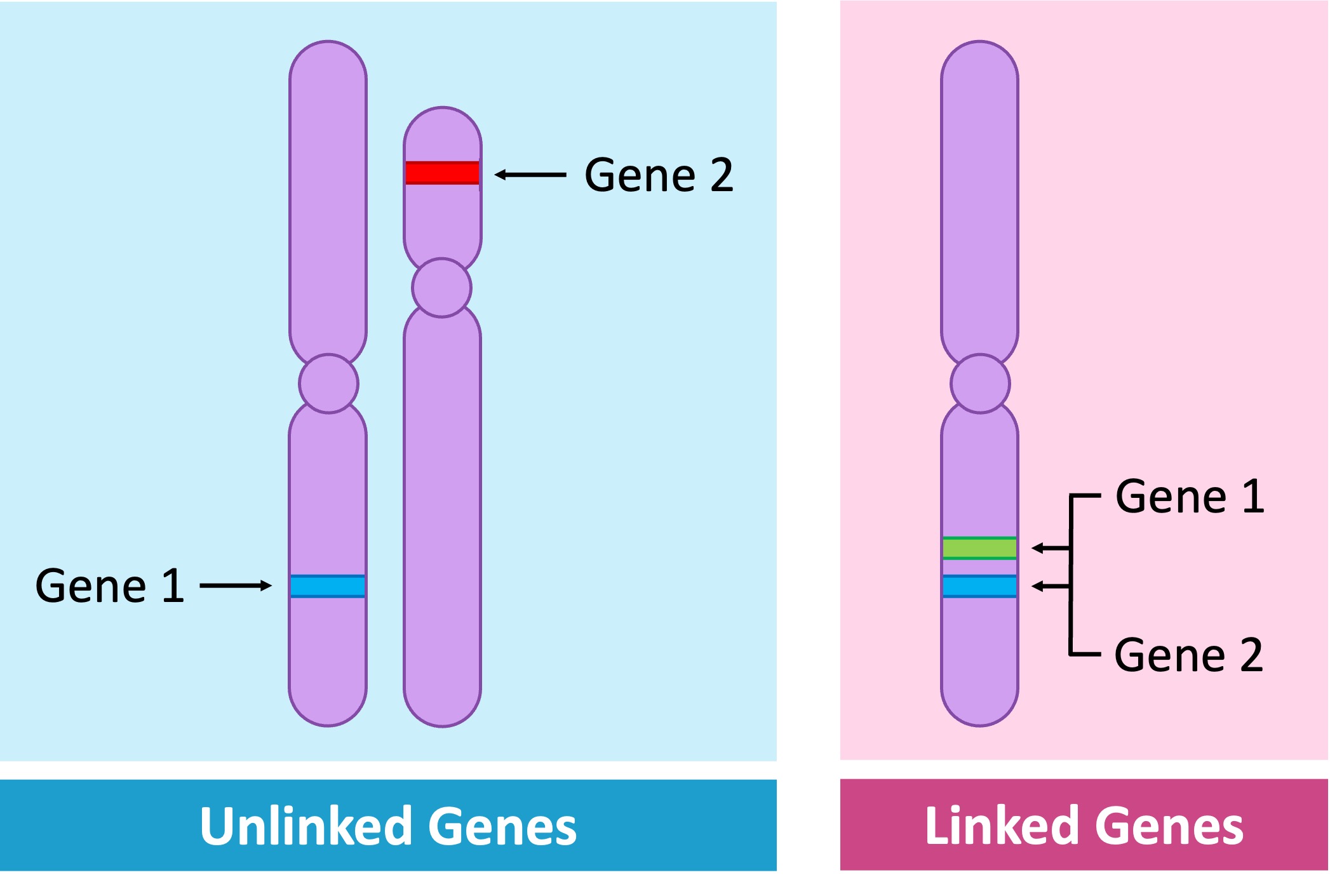
Linked Genes
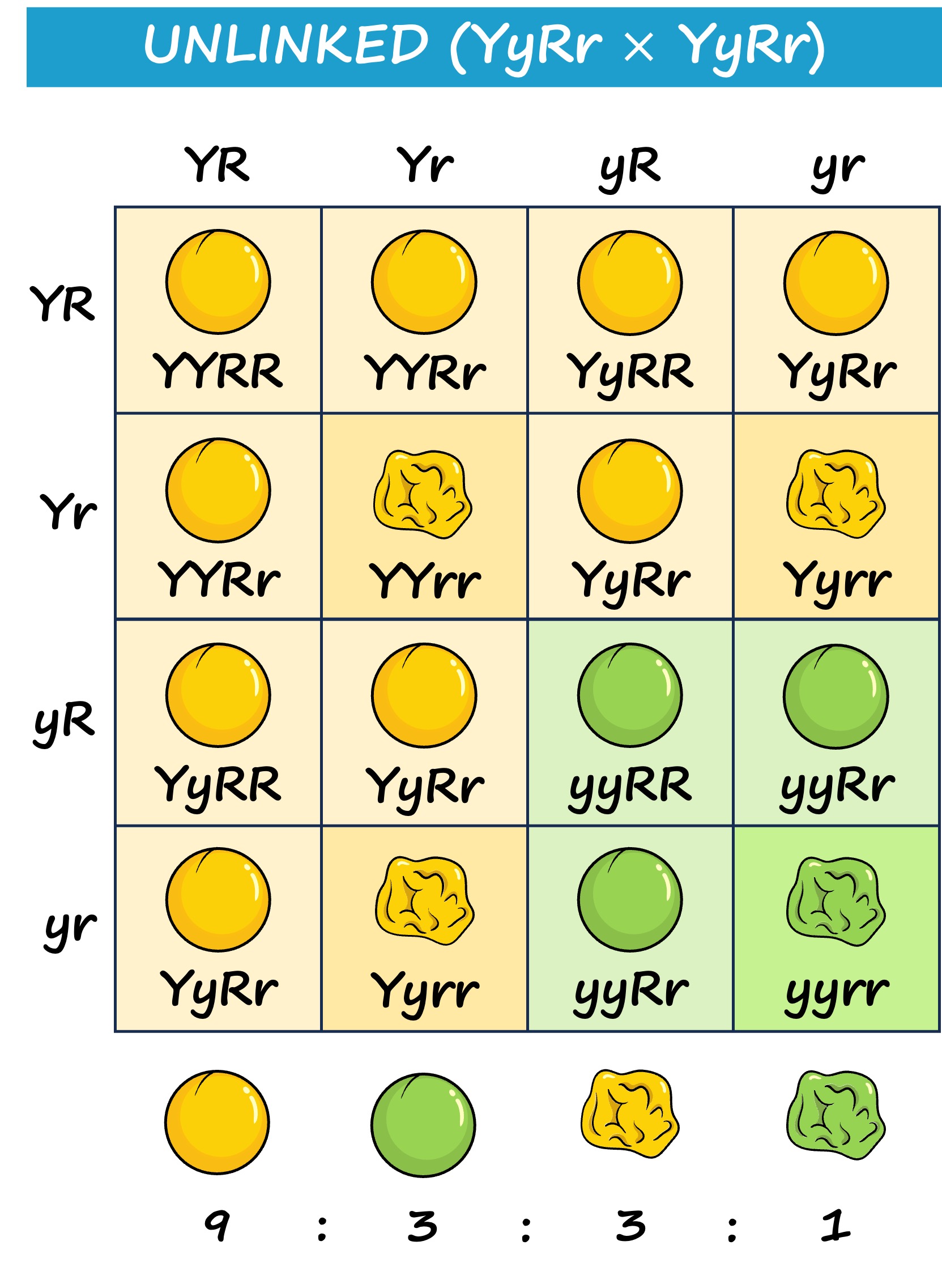
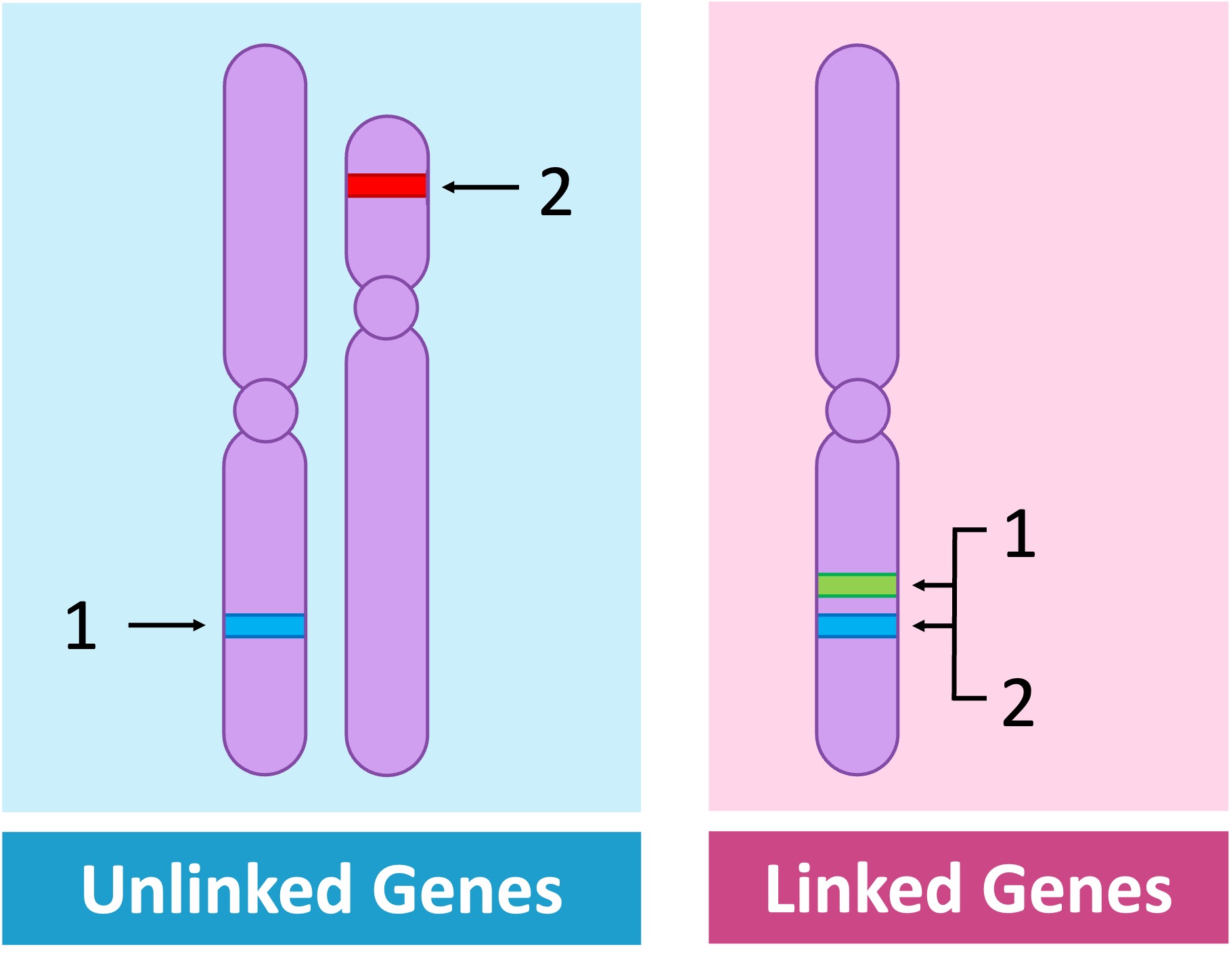
Unlinked genes are located on different chromosomes and will consequently demonstrate independent assortment
-
The specific position of a gene on a chromosome is called the gene locus (plural = loci)
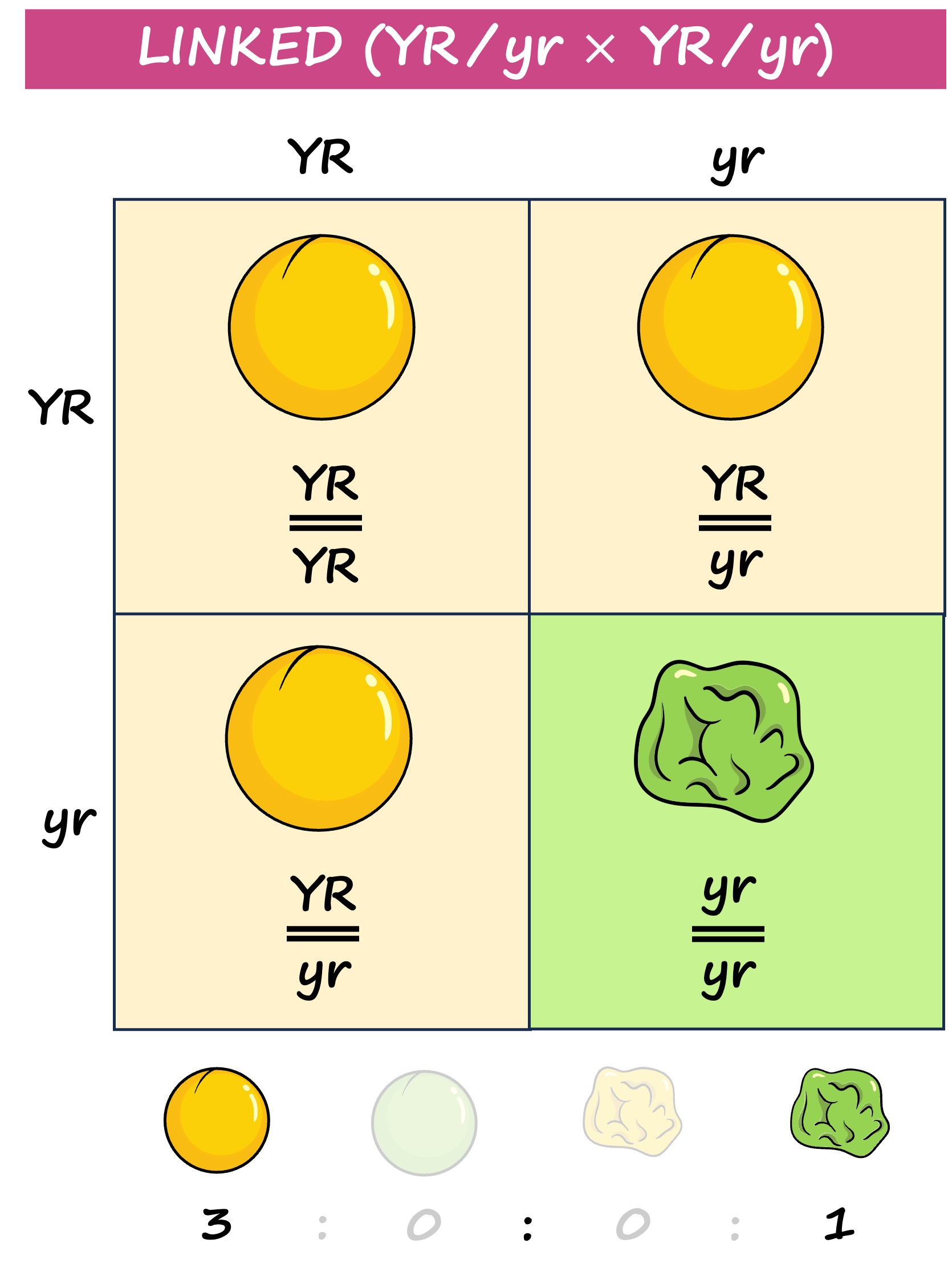
Linked genes are located on the same chromosome and hence do not demonstrate independent assortment
-
Linked genes are represented as vertical combinations – alleles pairs are shown alongside lines representing the homologous chromosomes (e.g. AB // ab)
The specific position of a gene (i.e. the locus) can be identified via three points of reference
-
The first point of reference is a number (or letter) which denotes the chromosome (e.g. 7q31 refers to chromosome 7)
-
The second point of reference is a letter (p or q) to denote which arm the locus is positioned on (e.g. 7q31 is on the lower q arm)
-
The third point of reference is a number corresponding to the proximity to the centromere (e.g. 7q31 is at a longitudinal position 31)
The locus of a human gene and its polypeptide product can both be identified using certain online resources
-
GenBank and EMBL are two genetic database that serve as annotated collections of DNA sequences
-
These databases can be used to establish whether two genes are linked or unlinked
-
Note: Genes on the same chromosome may be considered unlinked if they are far enough apart for recombination rates to reach 50%)
-
Gene Loci
Autosomal Inheritance
The patterns of inheritance will differ according to whether genes are linked or unlinked
-
Linked genes will be inherited together (no independent assortment) and hence don’t follow the normal inheritance patterns for a dihybrid cross
-
Instead the phenotypic ratios will be more closely aligned to a monohybrid cross as the two genes are inherited as a single unit
Inheritance Patterns