Gene Editing
The CRISPR-Cas9 system functions naturally in bacteria to provide immunity against viral infections
-
When a virus infects a bacterial cell, snippets of viral DNA are pasted into the bacterial genome to form a CRISPR locus
-
These snippets act as genetic memory bank (CRISPR = ‘clustered regularly interspaced short palindromic repeats’)
-
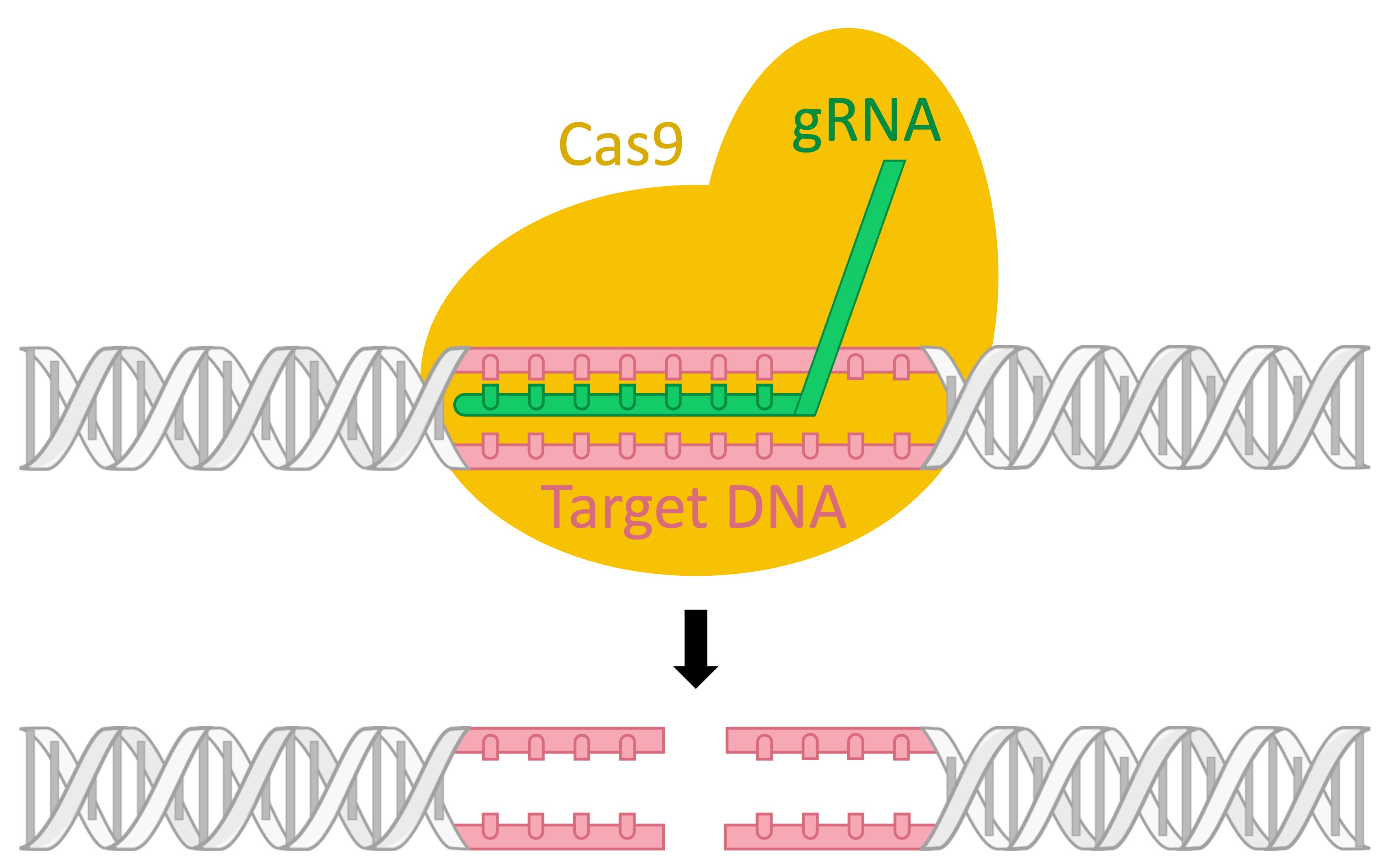
A CRISPR sequence is transcribed into a guide RNA strand (gRNA) that binds to a CRISPR-associated nuclease (Cas9)
-
The gRNA-Cas9 complex drifts throughout the cell until the gRNA locates and binds with any complementary viral DNA
-
This enables the Cas nuclease to then destroy the viral DNA sequence and hence prevents any subsequent infection
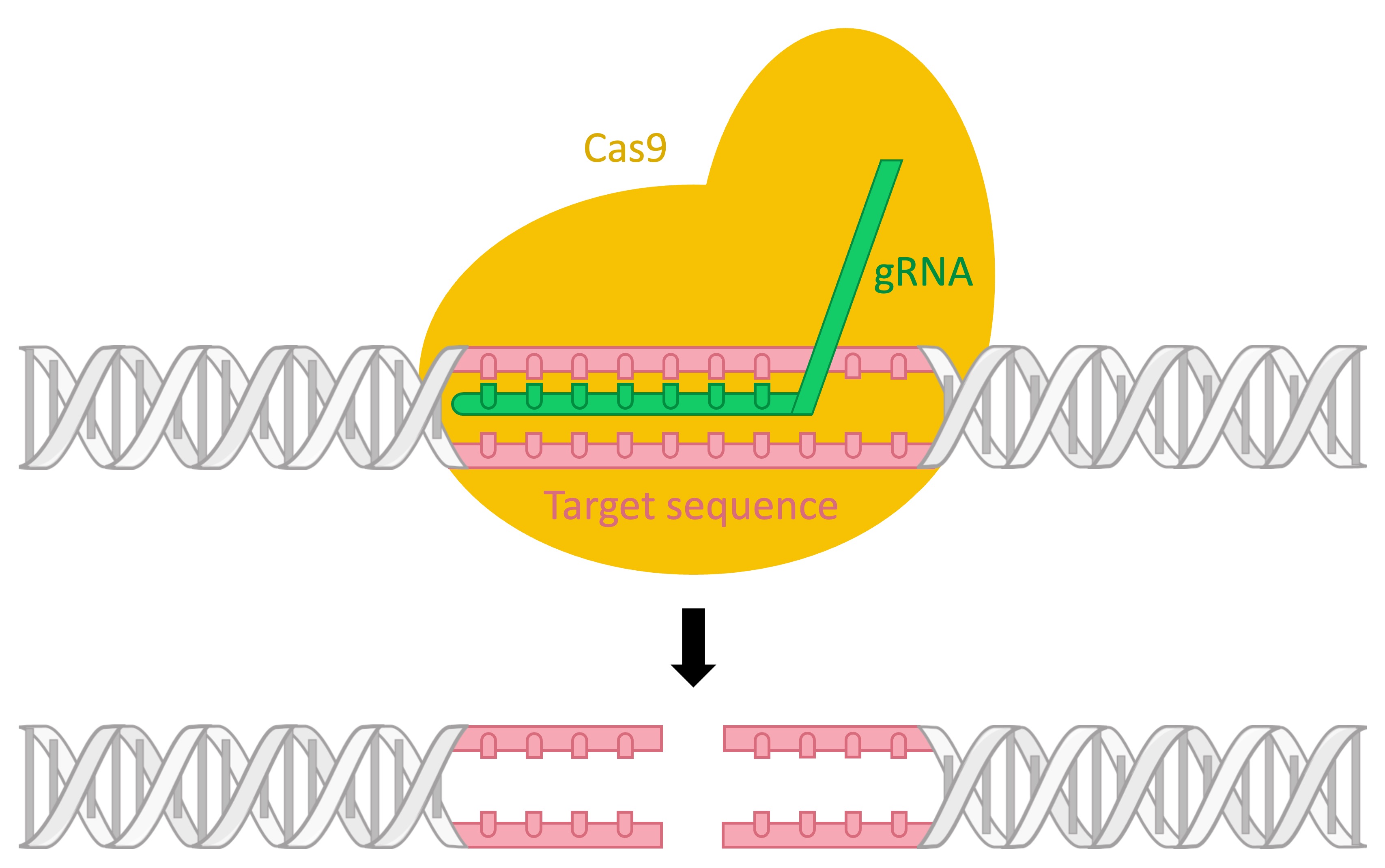
The CRISPR-Cas9 system has been modified by scientists to selectively remove any targeted sequence, allowing for precise gene editing
-
The Cas9 protein is complexed with a synthetically derived gRNA molecule that is complementary to a target sequence
-
The gRNA will bind to the target sequence, prompting its excision by the Cas nuclease (i.e. gene knockout)
-
Following the removal of the target sequence, another sequence of DNA can be integrated in its place (gene editing)
Gene editing via the CRISPR-Cas9 system has been used to address a variety of agricultural issues associated with food production
-
Certain metabolic pathways have been enhanced to improve nutritional content (e.g. higher starch production)
-
Plant absorption spectra have been modified to increase photosynthetic efficiencies (e.g. new pigments introduced)
-
Higher tolerances to biotic pathogens (viral, bacterial, fungal) or abiotic stresses (cold, drought, salt) have been achieved
-
Resistance to particular herbicides have been incorporated into crops to allow for elimination of competing weed species
CRISPR-Cas9 System