DNA Replication
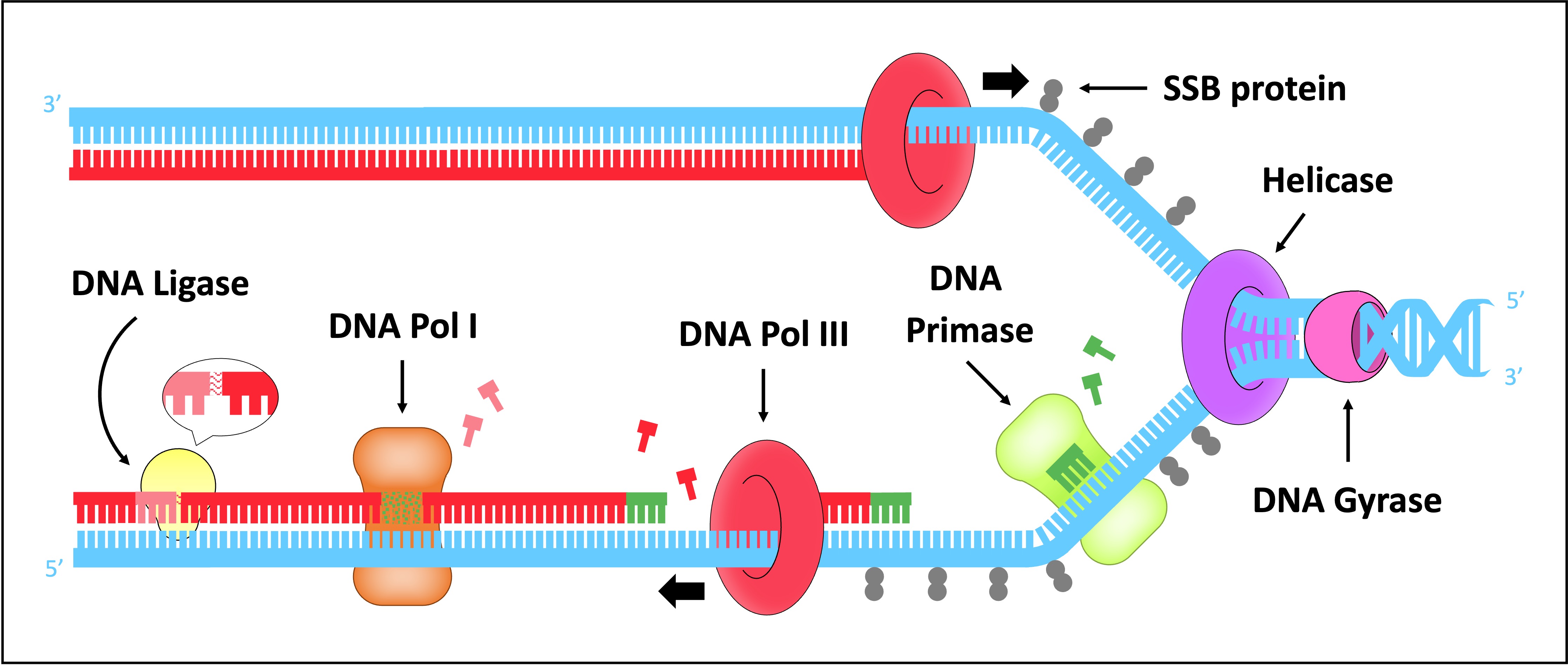
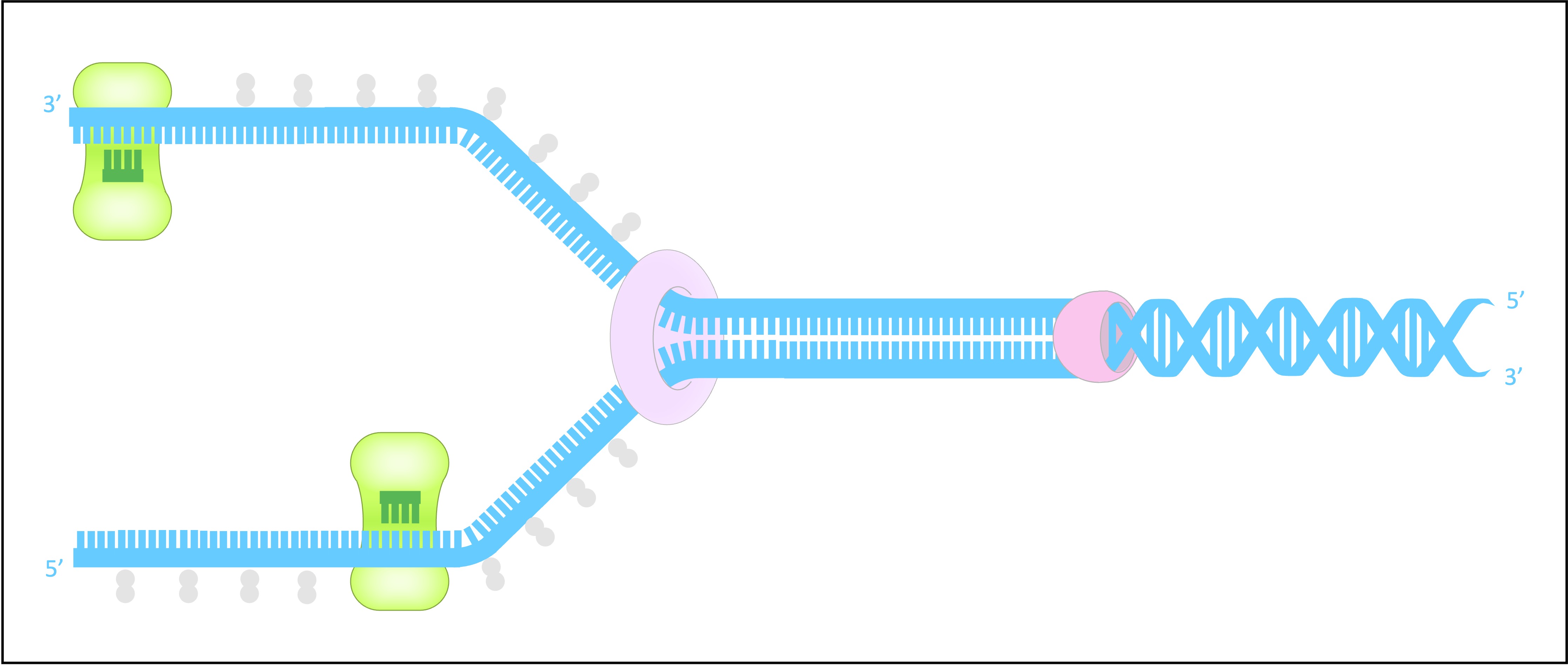
DNA replication is a semi-conservative process that is carried out by a complex system of enzymes
-
These include helicase, DNA gyrase, DNA primase, DNA polymerase (I and III) and DNA ligase
Helicase
-
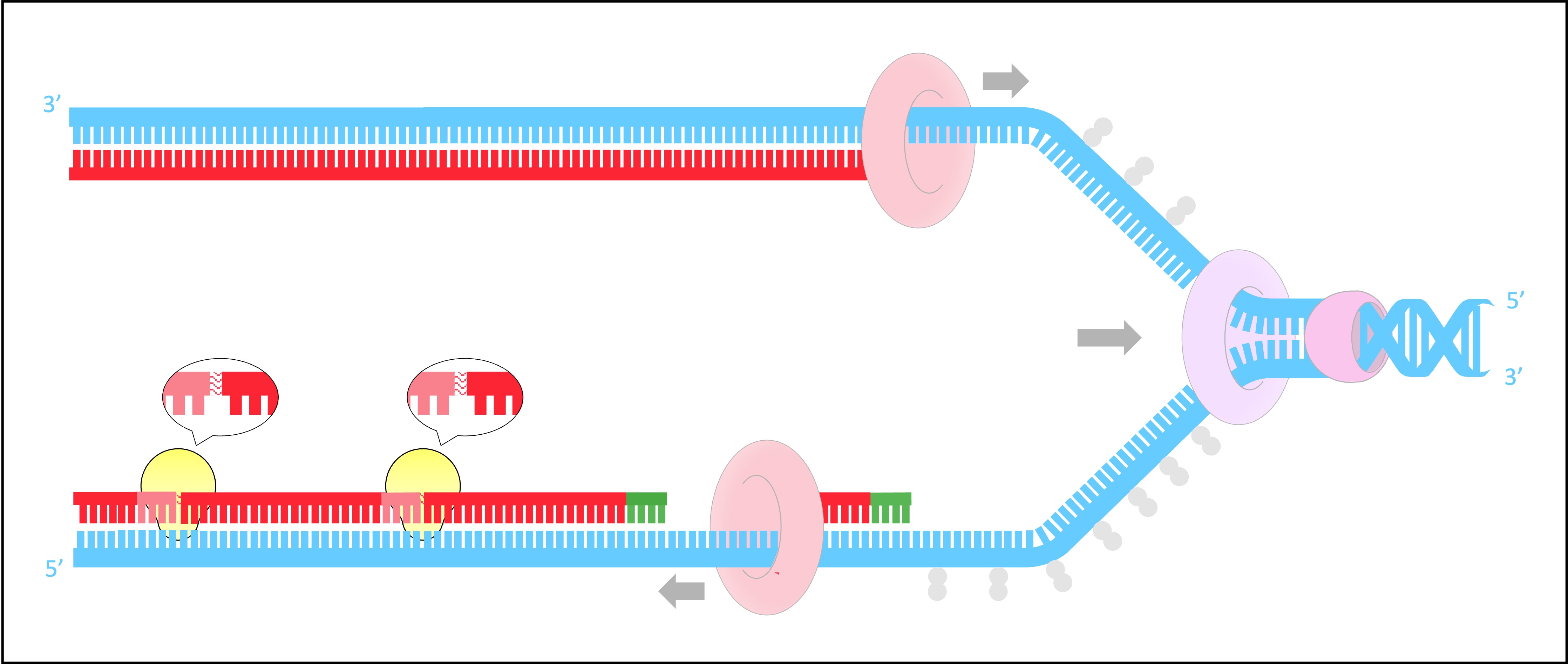
Helicase unwinds and separates the double-stranded DNA by breaking the hydrogen bonds between base pairs
-
This occurs at specific regions (origins of replication), creating a replication fork of two strands running in antiparallel directions
DNA Gyrase
-
DNA gyrase reduces the torsional strain created by the unwinding of DNA by helicase
-
It does this by relaxing positive supercoils (via negative supercoiling) that would otherwise form during the unwinding of DNA
Single Stranded Binding (SSB) Proteins
-
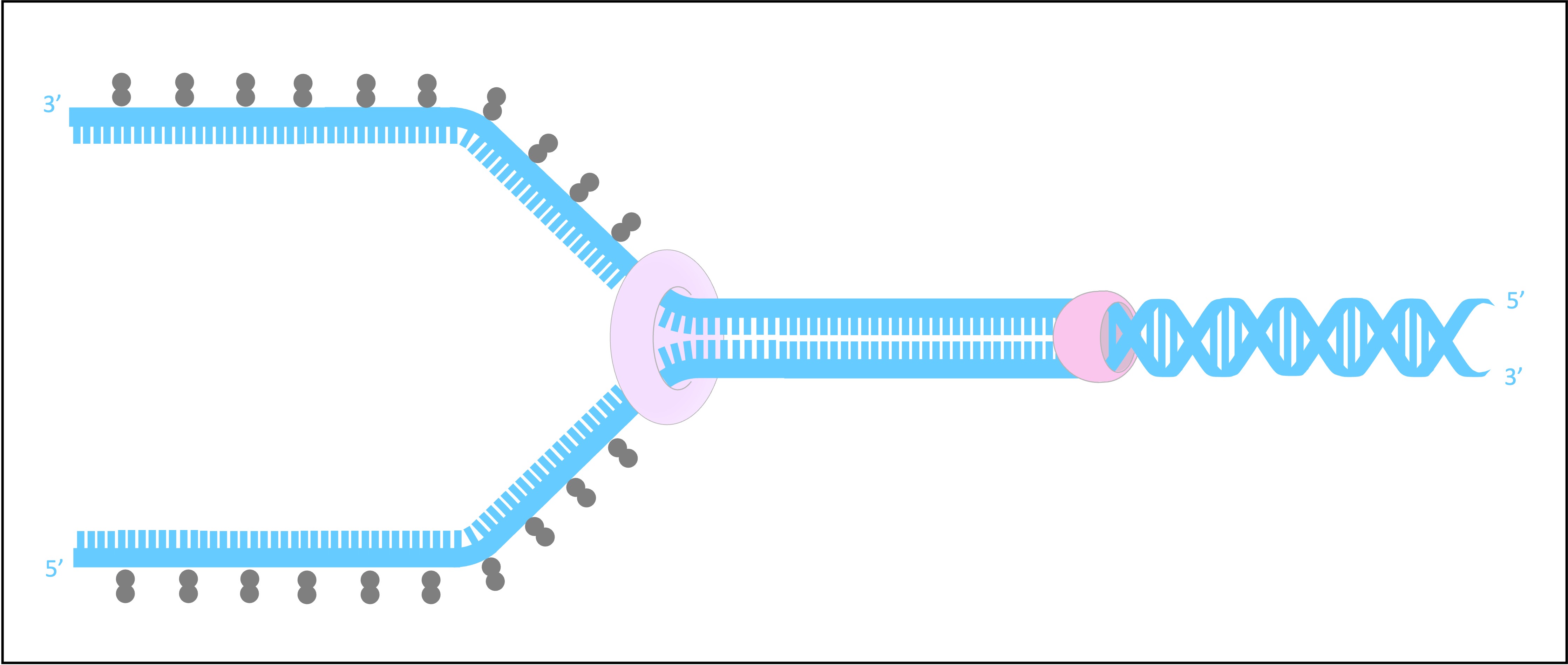
SSB proteins bind to the DNA strands after they have been separated and prevent the strands from re-annealing
-
These proteins also help to prevent the single stranded DNA from being digested by nucleases
-
SSB proteins will be dislodged from the strand when a new complementary strand is synthesised by DNA polymerase III
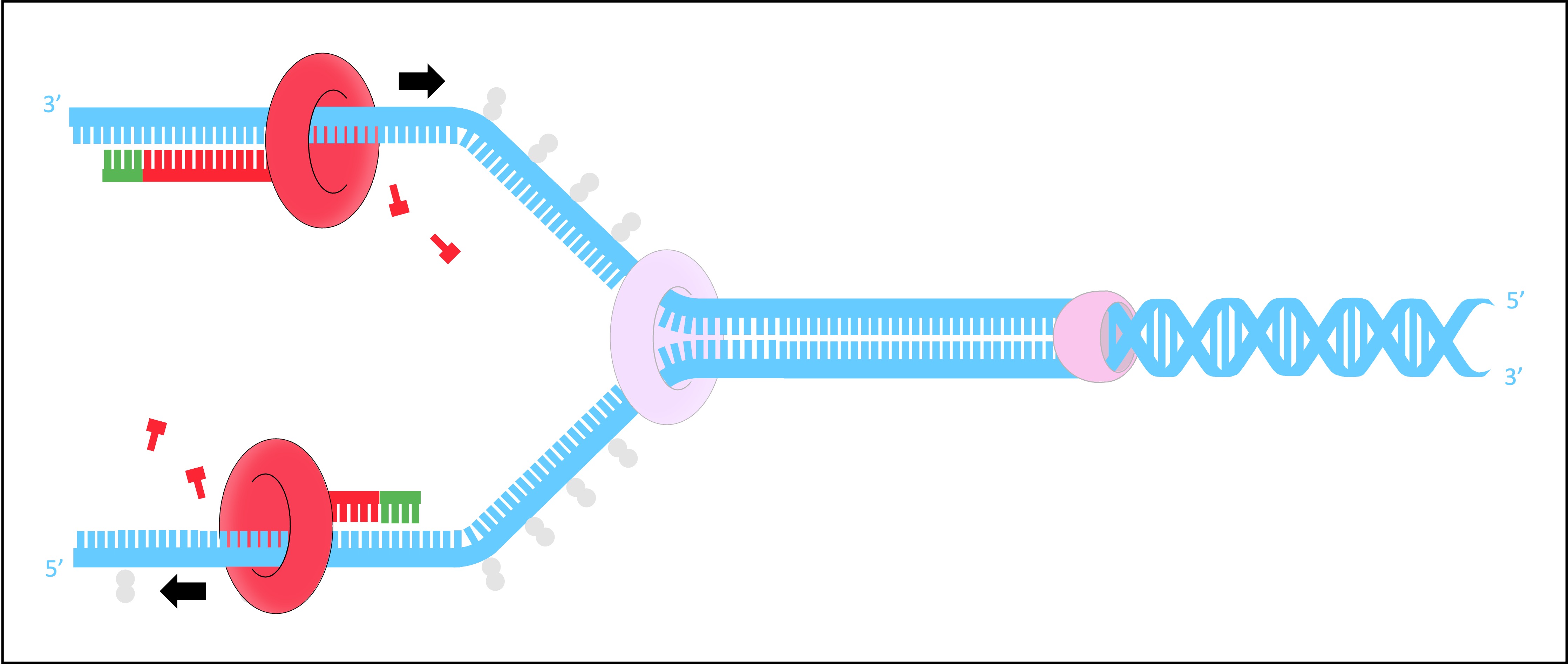
DNA Primase
-
DNA primase generates a short RNA primer (~10–15 nucleotides) on each of the template strands
-
The RNA primer provides an initiation point for DNA polymerase III, which can extend a nucleotide chain but not start one
DNA Polymerase III
-
Free nucleotides align opposite their complementary base partners (A = T ; G = C)
-
DNA pol III attaches to the 3’-end of the primer and covalently joins the free nucleotides together in a 5’ → 3’ direction
-
As DNA strands are antiparallel, DNA pol III moves in opposite directions on the two strands
-
On the leading strand, DNA pol III is moving towards the replication fork and can synthesise continuously
-
On the lagging strand, DNA pol III is moving away from the replication fork and synthesises in pieces (Okazaki fragments)
-
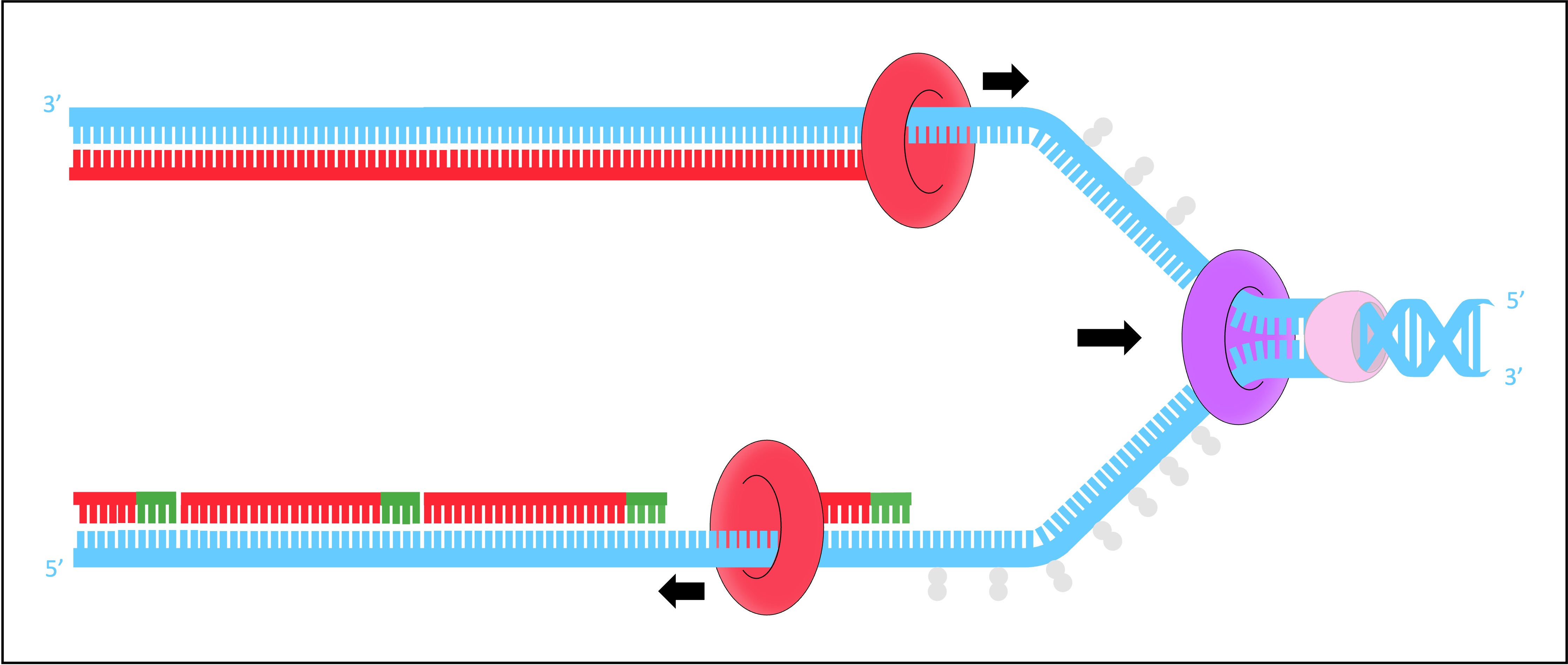
DNA Polymerase I
-
As the lagging strand is synthesised in a series of short fragments, it has multiple RNA primers along its length
-
DNA pol I removes the RNA primers from the lagging strand and replaces them with DNA nucleotides
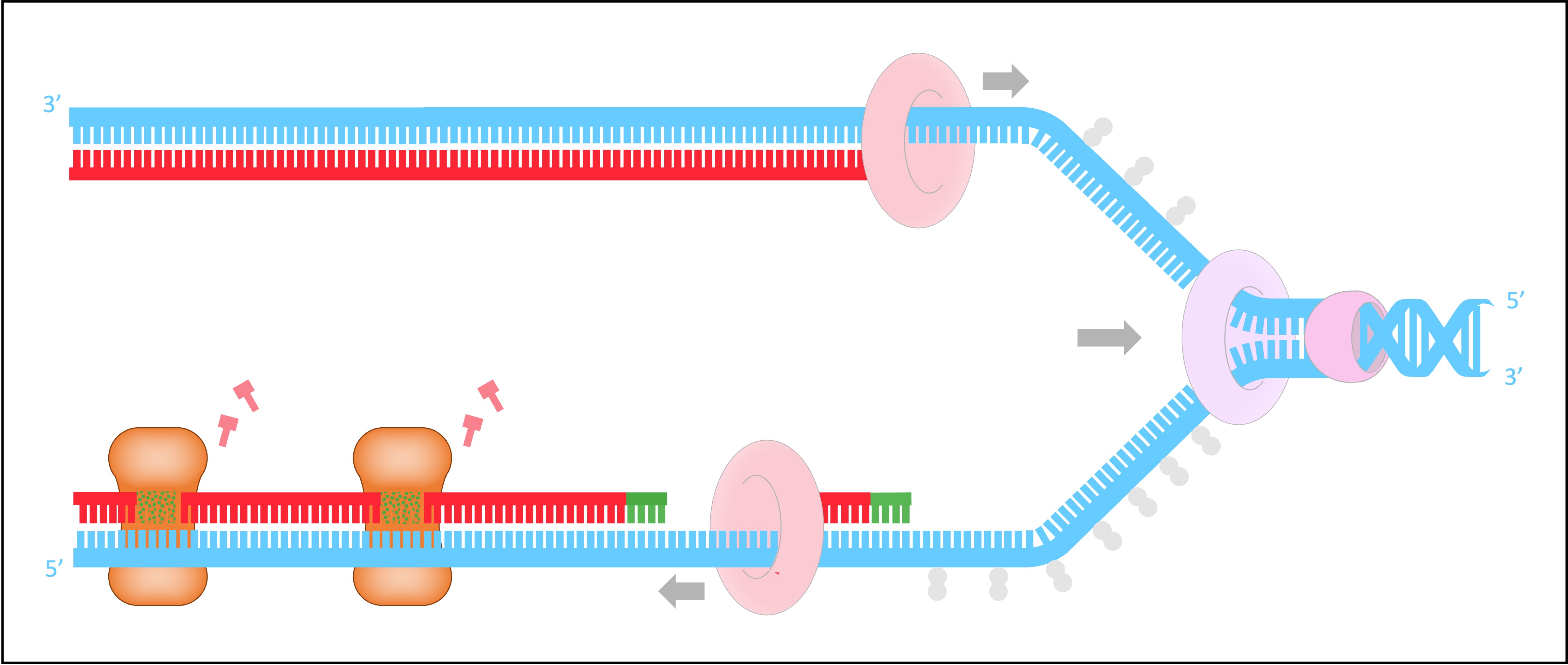
DNA Ligase
-
DNA ligase joins the Okazaki fragments together to form a continuous strand
-
It does this by covalently joining the sugar-phosphate backbones together with a phosphodiester bond